2. Power China Eco-Environment Group Co., Ltd., Shenzhen 518101, China
Over the past few years, organic solid waste has been extensively used because of its broad availability, large reserves, and diverse utilization forms[1]. The widespread reliance on fossil fuels has led to environmental damage and raised concerns over energy security. For this reason, a sustainable circular bio-economy is gradually developing, making large amounts of organic solid waste a potential feedstock for producing clean energy[2]. Anaerobic digestion (AD) is a widely used technique for processing organic solid waste[3], which can produce bio-methane, a form of clean energy[4]. To increase methane production during AD, various organic solid wastes are used for fermentation. The advantages of multi-substrate in AD include balancing nutrient content, improving digestion stability, enhancing methane production and digestion efficiency, and promoting a circular economy by utilizing various wastes[5]. Major organic substrates used in anaerobic co-digestion include animal manure, straw waste, sewage sludge, and so on. The ratios of co-substrates used in co-digestion processes differ significantly among various studies. For instance, in the co-digestion of poultry manure and wheat straw, the optimum condition is 55% of chicken manure in the substrate mixture[6]. In the study of Chen et al.[7], for the cattle manure to Chlorella sp. ratio of 8∶2, which can reach the higher methane production. Identifying the optimum substrate ratio, therefore, is essential for optimizing the benefits of AD for each substrate-type. However, these studies mainly focus on the co-digestion of two substrates, lacking research on the three or more substrates. To optimize the resource utilization of three or more organic solid waste[8-9], a better understanding of the material transformations, methane production rates, and methane yields during the AD is needed. However, the long AD period increases the time cost of experimental research[10-11]. Therefore, simulating the AD through theoretical or numerical modeling provides an effective approach to explore its optimization and control[12].
To monitor, optimize, and control AD, various mathematical models describing the process have been developed and documented[13]. The most famous and widely cited model is the ADM1 proposed by the IWA (International Water Association)[14]. The four-stage theory of anaerobic digestion, encompassing hydrolysis, acidogenesis, acetogenesis, and methanogenesis, forms the foundation of the model[15], which was initially developed to describe the anaerobic digestion of wastewater sludge, but it has now become the most commonly used structured anaerobic digestion model[16]. Due to its structured design, it is able to adapt to anaerobic digestion of various organic wastes, and this flexibility makes it perform excellently in multiple forms of anaerobic digestion[17-18]. For instance, the modified ADM1 can be used to predict the behavior of volatile fatty acid production in anaerobic digesters under varying operating conditions[19]. In the research of Sun et al.[20], the ADM1 modifications facilitate the evaluation of the effects of ammonia inhibition on methanogenic performance for common substrates, including acetate, formate, H2/CO2, and glucose. However, there is currently no clearly modified ADM1 model for AD of multiple organic solid waste. These studies overlooked substrate properties, which are crucial in the anaerobic digestion of various organic solid wastes, resulting in a lack of recommended kinetic parameters when simulating ADM1 for AD of multiple organic solid waste. By applying ADM1, precise process control and optimization of the anaerobic digestion process can be achieved. Some studies have shown that using ADM1 to simulate multi-substrate anaerobic digestion can optimize reactor operating conditions in subsequent experiments, effectively increase methane production, and reduce uncertainty and fluctuations in the process[21]. Additionally, the anaerobic digestion process of multiple substrates is complex, involving the degradation and gas production reactions of various organic components. ADM1 can provide a deep understanding of these complex processes, helping to address process challenges such as varying degradation rates and gas production efficiencies of different materials[22].
The main parameters related to methane production in the ADM1 model are influenced by experimental data, and the activity of functional microorganisms (such as methanogens) is the main factor affecting methane production. Therefore, functional microorganisms affect model parameters by influencing experimental data, which in turn affects the accuracy of model simulations. ADM1 does not include the microbial structure[23]. In this case, the analysis of community structure can determine the composition and changes of functional microorganisms during anaerobic digestion, which can lead to a better understanding of the simulation results of the model.
This study aims to investigate the effect of varying ratios of multiple substrates on methane production through a combination of experimental and simulation methods. A modified ADM1 model is proposed to accurately simulate the methane production capacity of various substrate ratios during AD. Additionally, from the perspective of the microbial community structure, the rationality of the model construction and the accuracy of the model simulation were further understood. This process not only deepens the understanding of anaerobic digestion mechanisms but also provides a theoretical basis and support for further operation and optimization of the process. The research results will help improve the efficiency of anaerobic digestion technology, promote the development of renewable energy, improve waste management strategies, and achieve a mutually beneficial situation for both environmental and economic gains.
List of abbreviations:
ac: acetic acid
kmi: The max specific degradation rate coefficient of i
ksi: The semi saturation coefficient of i
Si: Soluble component i
Xi: Microorganisms that degrade i
1 Materials and Methods 1.1 Feedstock and InoculumThe organic solid wastes used in this study include chicken manure (CM), corn straw (CS), vegetable waste (VW), reed (Re), and blue-green algae (Al). Among them, CM and CS were provided by a biogas plant located at Lan Kao in Henan Province, China. Reed was sourced from Xiong'an in Hebei, China. Al was obtained from Tai Lake in Jiangsu Province, China, and VW came from Laixi in Shandong Province, China. Al was subjected to dehydration treatment. CS, Re, and VW were dehydrated, ground, and filtered through a 20-mesh screen, then sealed and stored. The inoculum used in the experiments consisted of anaerobic sludge from a mesophilic lab-scale continuously stirred tank reactor that processed cow manure at Harbin Institute of Technology, Harbin, China. The basic characteristics of the materials and inoculum are outlined in Table 1.
| Table 1 Characteristics of substrates and inoculum |
1.2 Batch Experiments
Batch experiments were conducted in 250 mL glass bottles with screw caps, with a working volume set to 180 mL. The total solids content of the digested slurry was adjusted to 30% on a dry weight basis, with the inoculum ratio set to 10%. Following the addition of substrates and inoculum, to establish an anaerobic environment before fermentation, purging with N2 for 5 min to maintain an anaerobic environment. The fermentation temperature and initial pH were 37 ℃ and 7.0, respectively. Throughout fermentation, each reactor was manually shaken three times daily to ensure proper mixing. Three replicates were conducted in this study, and methane production was monitored daily. The mixing ratios of experimental group used for modifying ADM1 and conducting model validation are shown in Table 2.
| Table 2 The substrates mixing ratio for batch experiments to modify ADM1 |
1.3 Experimental Analysis
Total solids and volatile solids were measured according to standard procedures[24]. Methane production was determined using an Agilent 7890B gas chromatograph. Prior to measuring soluble indicators, samples were centrifuged at 12000 r/min for 5 min, and the supernatant was passed through a 0.45 μm filter membrane. The testing methods and parameters for volatile fatty acids (VFAs), such as acetic, propionic, iso-butyric, n-butyric, isovaleric, and n-valeric acids, were detailed in Tampio et al.'s study[25].
1.4 16S rRNA Gene Sequencing AnalysisThe microbial communities of samples from batch experiments were explored by Illumina Hiseq sequencing platform (Sangon Biotech, Inc., Shanghai, China). This study involved sampling from reactors to investigate potential changes in the microbial community composition, particularly the relative abundance of archaeal and bacterial populations. Relative abundance was determined as the ratio of sequences assigned to a specific taxon to the total number of sequences in each sample[26]. Genera representing less than 1% of the total composition in all samples were combined into a category labeled "others".
1.5 Establishment of the Model 1.5.1 Model modificationsIn this study, ADM1 was established using the AQUASIM 2.0, and simulations were carried out according to the methodology and initial values suggested by Batstone et al.[14]. The initial ADM1 model, depicted in Fig. 1(a), is grounded in the anaerobic digestion of sewage sludge. In a CSTR reactor, the model incorporates a composite particle substrate known as Xc, outlining the anaerobic digestion processes for Xc, which include disintegration, hydrolysis, acidification, acetification, methanogenesis, and microbial decay. Seven micro-organism types are involved in these processes, excluding the stages of disintegration and hydrolysis[27]. Research has proven that this model reliably represents the anaerobic digestion associated with sewage sludge[28]. However, the original ADM1 model is only applicable to anaerobic digestion of a single substrate such as sludge, and cannot fully describe the synergistic effects between multiple organic solid wastes (OSWs). These synergistic effects include the impact of substrate diversity on microbial communities, the promotion of system stability by nutrient balance between different substrates, and the regulation of the generation of different volatile fatty acids, all of which directly affect methane production. However, the unified substrate Xc used in the original ADM1 model cannot accurately simulate the complex interactions of these diverse substrates. Therefore, this study modified the structure of the ADM1 model to better describe the dynamic processes in multi substrate anaerobic digestion. The uniform Xc in the original model is used to represent the organic substances in the reactor, but cannot simultaneously describe the substrate (OSW) and inoculum (biogas slurry) with significant differences.
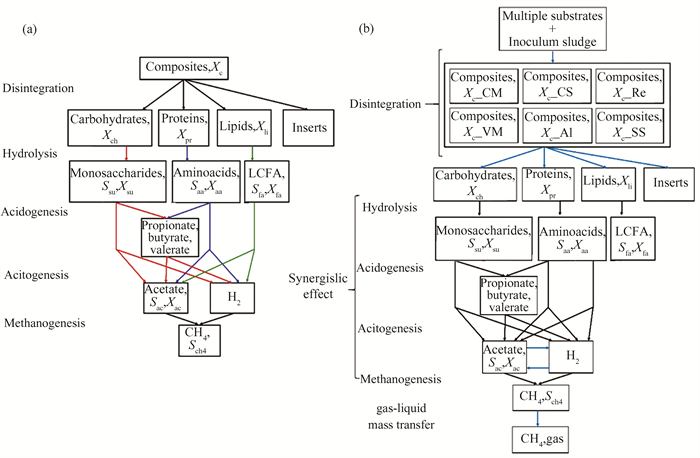
|
Fig.1 (a) Initial ADM1 model and (b) modifed ADM1 model |
Compared with sewage sludge, multiple OSW are more granular and have higher levels of carbohydrates and fats. Meanwhile, these substrates have a synergistic effect on each other. The synergistic effect is manifested in the following aspects. By mixing different matrices, the C/N can be adjusted to make microbial growth more efficient; The diversity of microbial communities supported by different substrates improves the stability and degradation efficiency of the system; Balance different types of VFAs, avoid system acidification, and promote methane generation; Dilute toxic substances such as ammonia nitrogen and algal toxins to reduce their negative impact on microorganisms. Overall, these synergistic effects collectively enhance the overall efficiency and stability of the system, increasing methane production. To accurately simulate the AD of OSW, it is essential to consider the unique characteristics of OSW separately and sludge. In this study, modifications were made to the original ADM1 structure to incorporate processes specific to OSW. The revised model structure is shown in Fig. 1(b). Unlike the original ADM1, the proposed model distinctly addresses different substrates, including biogas slurry and organic solid waste, as well as their interactions.
Following the methodology of Zhao et al.[27], the kinetics and rate-based equations of the starting ADM1 model were adjusted to align with the revised model structure. During the hydrolysis and acidification stages in AD of multiple OSW, similar small-molecule compounds like monosaccharides, VFAs, and others are produced. However, significant differences exist in the disintegration stages of sludge and OSW.
In this work, OSW exhibits significant synergistic interactions, unlike sludge, which is a key reason for modifying the initial ADM1. The cooperation between substrates is essential when accurately describing the degradation of complex OSW. The degradation of complex OSW typically follows a surface-based kinetic model, emphasizing the need to precisely depict the decomposition of particulate OSW by microbes. Therefore, the disintegration process (ρ1) from the original ADM1 was divided into several parts: one pertains to biogas slurry (ρ1-1) and the rest pertain to OSW. The disintegration rate equation for biogas slurry remains unchanged. Table 3 presents the matrix representing the equations for these alterations to the model, whereas the unchanged parts of the model are detailed in the work by Batstone et al[14].
| Table 3 Matrix of modified ADM1 equations |
1.5.2 Modification of state variables
Values for the substrate state variables within the model were established through the experiments. In particular, Xc_CM, Xc_CS, and other variables denote the initial quantities of OSW introduced into the reactor. The initial quantities were determined following the design principles of the batch experiments, with all measurements expressed as COD. The stoichiometric parameters fch_Xc, fli_Xc, fpr_Xc, and fxi_Xc were calculated according to the method outlined by Zhao et al.[27], as depicted in the Eq.(1):
| $ \begin{aligned} & f_{\mathrm{ch}_{-} X \mathrm{ci}}=\frac{\mathrm{CH}_i}{\mathrm{VS}}, f_{\mathrm{pr}_{-} X \mathrm{c}}=\frac{\mathrm{PR}_i}{\mathrm{VS}}, \\ & f_{\mathrm{li}_{-} X_{\mathrm{c}}}=\frac{\mathrm{LI}_i}{\mathrm{VS}}, f_{\mathrm{xi}_{-} X \mathrm{c}}=\frac{I_i}{\mathrm{VS}} \end{aligned} $ | (1) |
where CHi, PRi, PLi and Ii(i=1, 2, 3, 4, 5)denote the carbohydrates, proteins, lipids, and inert compounds present in OSW. The other model components remain unchanged from the original ADM1, and thus the stoichiometric and kinetic parameters follow the recommended values. Modifications and simulations of the model were conducted using AQUASIM 2.0 software.
1.5.3 Sensitivity analysis and parameter estimationA sensitivity analysis was conducted to identify the ADM1 parameters that most significantly influence methane production. The parameters examined included the disintegration constant of composite particulates (kdis), the maximum specific substrate uptake rates and half-saturation constants as defined by Monod's kinetics. Methane yield was used as the output metric for the sensitivity analysis due to its importance to the study's objectives. The formula for calculating the sensitivity index (SI) is presented in Eq.(2):
| $ \mathrm{SI}=\sigma_{q, p}^{a, a}=\frac{\partial q}{\partial p} \cdot \frac{p}{q} $ | (2) |
where q represents the target parameter, and p represents the analysis parameter. The sensitivity index (SI) indicates the absolute change in q when p changes by 100%. When the SI is positive, it indicates a positive correlation between p and q; conversely, a negative SI indicates a negative correlation.
1.5.4 Model evaluationIn this study, the goodness of fit R2 is used as an important metric for model evaluation. This method is commonly employed to assess how well the experimental values match the simulated values. The calculation formula is provided in Eq. (3):
| $ R^2=1-\left(\frac{\sum(y-z)^2}{\sum y^2}\right)^{0.5} $ | (3) |
where y denotes the experimental data as well as z denotes the simulated values. The value of R2 lies between 0 and 1, with greater values representing a better fit. In this work, the goodness of fit is used to analyze how well the simulated values match the experimental values, aiming to obtain a model with a high goodness of fit.
2 Results and Discussion 2.1 Batch Fermentation for Methane Production from Multiple OSWFig. 2(a) shows the methane production of control groups using a single substrate during 30 days of AD. Compared with the reactor that use CM, reactors that use plant-based OSW (straw and blue-green algae) produced less methane.
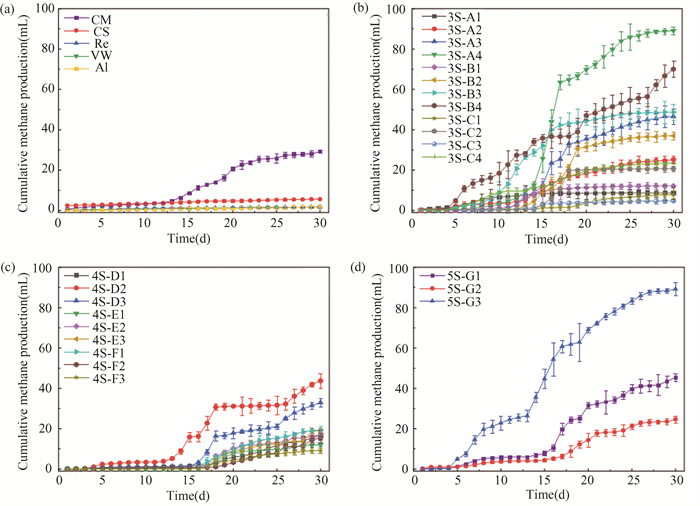
|
Fig.2 Methane production in (a) one substrate (b) three substrates (c) four substrates (d) five substrates experimental batch reactors |
In Fig. 3, the microbial analysis revealed that Firmicutes(51.12%), Bacteroidota (39.95%), Proteobacteria (5.03%) and Synergistota (1.08%) were the predominant phyla in the CM used. These phyla are frequently identified in the anaerobic digestion process, as they can break down carbohydrates and proteins and play a crucial role in the breakdown of VFAs.

|
Fig.3 The relative abundance of the phyla Firmicutes, Bacteroidota, Proteobacteria, and Synergistota at the class, order, and family levels |
The phylum Firmicutes (Fig. 3) was mainly represented by the taxonomic class Bacilli, which constituted45.08% of the phylum. Within Bacilli, the orders Bacillales and Lactobacillales accounted for 34.56% and 8.60% of the phylum, respectively. This group is commonly found in the digestive or gastrointestinal tracts of animals and is mainly responsible for the hydrolysis of plant polysaccharides and the transformation of carbohydrates (such as fructose, maltose, lactose, and galactose) into acetate, ethanol, or a mixture of VFAs under anaerobic conditions[29].
The phylum Bacteroidetes is primarily composed of the order Bacteroidia (Fig. 3). Species within the phylum Bacteroidetes are renowned for their role in protein degradation. Most proteolytic microorganisms can metabolize amino acids, resulting in the production of NH3 and VFAs such as acetate, propionate, and succinate. As for the phylum Proteobacteria, it is characterized by the taxonomic class Gammaproteobacteria, which makes up 4.38% of the phylum.
As for the phylum Synergistota, they participate in the primary degradation of organic matter, but also contribute to maintaining the micro-ecological balance of the anaerobic fermentation system through their metabolic activities, regulating the pH and other environmental parameters to ensure process stability. These taxonomic findings indicate that the dominant phyla in poultry manure are involved in the breakdown of complex organic substances like proteins and carbohydrates, which are then fermented into acetic, propionic, butyric, and iso-valeric acids as the primary products.
Therefore, in a single-substrate AD system, the reactor using CM yield better methane production compared with those using straw-like substrates. In multi-substrate AD systems, the addition of CM increases methane production.
2.2 Model Calibration 2.2.1 Sensitivity analysisThe results of the analysis of sensitivity index are shown in Fig. 4. The decay rate constant for microbes (kdec) showed the highest SI values, with a gradual upward trend peaking at 0.36. The substrate disintegration rate constant (kdis) also exhibited significant SI values, peaking at 0.26 at 20 days and then falling rapidly to 0.011 as the reaction finished. High SI values were also observed for the parameters km_ac and KS_ac. These play a crucial role in determining the rate of methane production, representing the methanogens' maximum substrate utilization rate and substrate affinity, respectively. Specifically, km_ac is the Monod maximum specific uptake rate for acetic acid, while KS_ac is the half-saturation constant for acetic acid. The sensitivity index of km_ac reaches its peak in the later stage of the reaction and drops to zero after 20 days. In contrast, the sensitivity of KS_ac shows the opposite trend, gradually decreasing as the reaction progresses. These changes indicate that the substrate utilization rate and half saturation constant have different effects at different stages of methane generation. Compared with Zhao et al.'s study [27], the peak of km_ac in this study appeared in the later stage of the reaction, mainly due to differences in substrate types. Zhao et al. used a single food waste as a substrate, while this study used multiple substrates including CM and CS. The different degradation characteristics of these substrates result in different lag periods, thereby affecting the sensitivity changes of km_ac and KS_ac. In addition, the mixed use of multiple substrates can adjust the C/N ratio, thereby having different effects on microbial activity and substrate degradation rate, which are reflected in the changes in kinetic parameters.
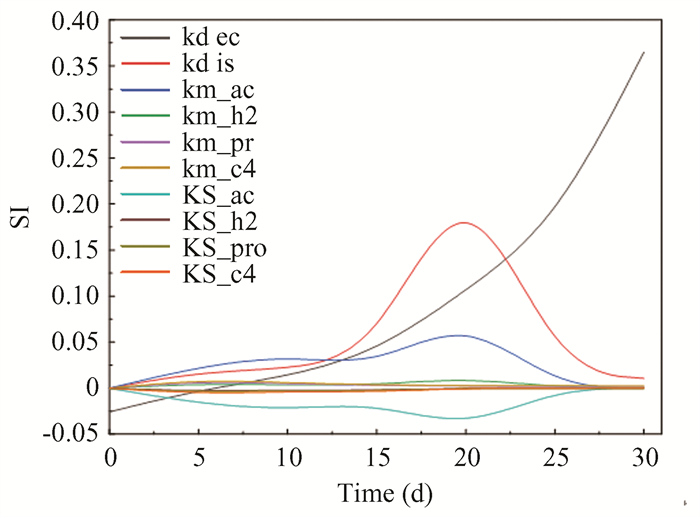
|
Fig.4 SI analysis of kdec, kdis, km_ac, km_h2, km_pr, km_c4, KS_ac, KS_h2, KS_pro, and KS_c4 |
In addition to kdec, kdis, km_ac, and KS_ac, the sensitivity changes of other parameters such as km_h2, km_pro, km_c4, and KS_h2 also have a significant impact on the methane generation process. For example, km_h2 showed high sensitivity in the early stages of the reaction, indicating that the rate of hydrogen generation has a significant impact on methane production, while km_c4 showed high sensitivity in the later stages of the reaction, reflecting the contribution of butyric acid degradation rate. Compared with these positively sensitive parameters, KS_h2 and KS_pro exhibit negative values, indicating the limiting effect of the semi saturation constants of these substrates on the reaction rate. In addition, the changes in these parameters are closely related to the differences in microbial activity and substrate types at each stage.
2.2.2 Parameter estimationThe four parameters identified as sensitive, kdec, kdis, km_ac, and KS_ac, were estimated using experimental results. Initially, the simplex method was applied, followed by the secant algorithm to enhance convergence. Both algorithms are integrated into AQUASIM 2.0 software. Based on the sensitivity analysis results, the model parameters listed in Table 4 were adjusted to calibrate the ADM1 model in this study.
| Table 4 Parameter values and R2 of calibration experimental values |
Batstone et al.[14] found that the parameters of the ADM1 model are mainly influenced by reaction conditions. Therefore, parameter estimation was recalibrated according to the sensitivity analysis and experimental findings. Among the calibration parameters, the adjusted kdec value showed significant deviation from its starting value. The optimal kdec value for the anaerobic digestion of OSW was found to be 0.03, based on the average of optimized kdec values obtained during the tuning experiments. This value is marginally higher than the originally recommended ADM1 value of 0.02.
The findings reveal that the anaerobic digestion system in this study enables microorganisms to degrade OSW at a faster rate compared with the initial sludge AD system. The calibrated kdis value exceeded the initial 0.5, which shows that the modified ADM1 model effectively considered OSW particle characteristics. Smaller particle sizes result in a larger specific surface area, improving interaction with microorganisms, as evidenced by the increased kdis parameter. In the model, km_ac and KS_ac are fundamental kinetic parameters involved in methanogenesis. The km_ac value was initially set at 8 but was later revised to 3.64, while KS_ac was adjusted from 0.15 to 0.27.
Additionally, Batstone et al.[14] also examined the anaerobic digestion of different substrates, including primary sludge, organic acids, and carbohydrates. Authors proposed that the calibration value ranges for km_ac and KS_ac be between 0.14 and 52, and between 0.011 and 0.93, respectively. The values proposed in the present study are within these recommended ranges.
2.2.3 Comparison of simulated results with experimental dataTable 4 shows that the average R2 values for the 3S, 4S, and 5S simulations with the initial ADM1 model were 0.814, reflecting a weak correlation between the simulated outputs and the experimental data for biogas production. Figs. 5 (a), (e), and (f) reveal a significant difference in the methane production trends produced by the initial ADM1 model compared with the experimental results. The kdis and KS_ac in the original ADM1 model failed to reflect the differences of different substrates, resulting in prediction bias in the degradation of multiple substrates. In addition, the original model did not fully consider the synergistic effects between substrates, such as the mutual influence between straw and chicken manure during the degradation process. These factors make it difficult for the model to accurately predict methane generation. By optimizing parameters such as kdis and KS_ac, and introducing new collaborative degradation pathways, the modified ADM1 model better simulates the actual process of multi substrate anaerobic digestion, significantly improving prediction accuracy. Similarly, Chen et al.[30] observed that the ADM1 model gave inaccurate biogas yield estimates from Hydrilla verticillata without parameter optimization.

|
Fig.5 Experimental methane production results of 3S-A, 3S-B and 3S-C compared to the modified ADM1 results of 3S-A, 3S-B and 3S-C |
Nonetheless, following parameter optimization, the discrepancy between the model predictions and the experimental data was significantly reduced(Figs. 5(b), (d), and (f)). Specifically, the R2 values for the 3S-A, 3S-B, and 3S-C groups improved to 0.977, 0.972, and 0.968, respectively (Table 4). This indicates that the adjusted ADM1 model with tuned parameters can accurately forecast methane generation from OSW anaerobic digestion. Razaviarani et al.[31] observed that the revised ADM1 model effectively estimated methane yield levels in laboratory-scale anaerobic digestion systems, closely matching experimental results.
2.3 Model ValidationFig. 6 illustrates the fit degree between the results from the adjusted ADM1 simulation and the observed data for total methane yield. The values of R2 for the simulations were 0.960, 0.946, 0.976, and 0.977, respectively, as shown in Table 5, closely aligning with those from the calibration experiments. This demonstrates that the adjusted ADM1 is capable of reliably forecasting the results of batch experiments. At the start of fermentation, the reactor contains dissolved organic matter and natural alkalinity, providing ample substrates and an optimal environment for methanogenesis[32]. Initially, swift breakdown of the substrate generates significant quantities of VFAs, which suppress methanogen function and consequently lower methane yield. As the process of anaerobic digestion progresses and the organic matter content diminishes, the production of VFAs decreases due to substrate limitation. This reduction in VFAs alleviates the inhibition on methane production, leading to a gradual increase in methane yield. However, the modified ADM1 model does not incorporate microbial responses to environmental changes, resulting in simulations that reflect purely mechanical local phases[33]. Future research will aim to optimize the modified ADM1 to better account for these dynamics.

|
Fig.6 Methane production of (a) initial ADM1 simulation effect and (b) modified ADM1 simulation effect |
| Table 5 R2 of validation experimental data |
2.4 Microbial Community Analysis
Community structure is a key factor influencing cumulative methane production. Therefore, 16S rRNA gene sequencing technology was employed to analyze the community structure of both the methane production experimental groups and the control group. Fig. 7 (a) shows the distribution of methanogenic archaea at the genus level, while Fig. 7 (b) presents the bacteria associated with the methane production process at the genus level.
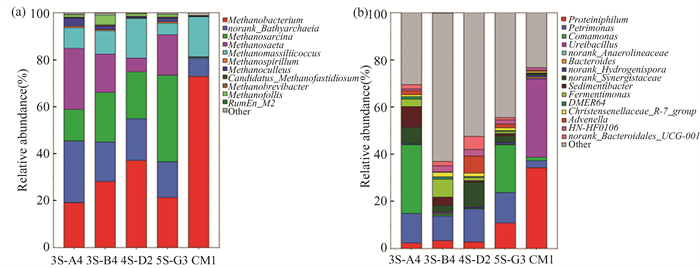
|
Fig.7 Variations of archaea(a) and bacterial(b) community composition at genus levels |
Within Fig. 7, in the control group(CM1), the dominant genera were Methanobacterium[34](72.98%) and Methanomassiliicoccus[35](17.08%). In the experimental groups, the dominant genera were Methanobacterium[36], norank_Bathyarchaeia[37], Methanosarcina[38], and Methanosaeta[34]. Among them, except for norank_Bathyarchaeia, the rest are methanogenic archaea.
As shown in Fig. 7(a), the archaeal composition and relative abundance in samples from reactors A4, B4, D2, and G3 were similar, with Methanosarcina and Methanosaeta being the dominant genera. Methanosarcina, a facultative methanogen that can metabolize both hydrogen and acetate, predominantly carried out acetate-utilizing methanogenesis in these reactors. It was the most abundant acetate-utilizing methanogen in reactors with high acetate concentrations[20]. The consistency between the 16S rRNA analysis results and the model simulation confirms the reliability of the modified ADM1 and the simulated key kinetic parameter values.
As depicted in Fig. 7(b), Petrimonas and Comamonas play a crucial role in the early degradation of organic matter, particularly by promoting the degradation of complex carbohydrates and generating intermediate products such as VFA. These intermediate products provide substrates for methane producing bacteria Methanosarcina and Methanosateta, further promoting methane production. Therefore, changes in bacterial community structure are closely related to the generation and transformation pathways of VFA, ultimately affecting methane production efficiency. The relative abundance of Comamonas in reactors 3S-4A and 5S-G3 was significantly higher than in reactors 3S-B4 and 4S-D2. Additionally, the higher relative abundance of norank_Synergistaceae in reactor 4S-D2 (10.45%), compared with other reactors (approximately 2.8%-6.70%), was consistent with the relatively higher presence of Methanobacterium (Fig. 5(a)).
The above results indicate that the trends in the changes of methane-producing microorganisms align with the trends in parameters such as km and KS estimated in the ADM1 model[27, 39]. This consistency suggests that the quantitative descriptions of the methane production pathways in the revised ADM1 model match the microbiological results. Therefore, this study demonstrates that the modified and extended ADM1 can be considered an effective tool for both macroscopic and microscopic evaluation of the anaerobic digestion process of multiple materials[40]. In other words, this model can not only simulate the gas production process during the experiment but also predict the changes in the abundance of methane-producing microorganisms throughout the experimental process.
3 ConclusionsThis study assessed the methanogenic performance of various organic solid wastes (chicken manure, corn straw, reed, vegetable waste, and blue-green algae) by modifying the ADM1 model and characterizing the microbial community. Key kinetic parameters (kdec, kdis, km_ac, and KS_ac) were calibrated because of their substantial influence on methane production, with suggested values of 0.03, 6.07, 3.64, and 0.27, respectively. Methanosarcina and Methanosaeta were identified as dominant methanogens.The microbial characterization and the modified ADM1 simulations corroborated each other. The modified ADM1 accurately predicts methane production from OSW under varying substrate characteristics and operational conditions, demonstrating its versatility.
| [1] |
Ma H, Guo Y, Qin Y, et al. Nutrient recovery technologies integrated with energy recovery by waste biomass anaerobic digestion. Bioresource Technology, 2018, 269: 520-531. DOI:10.1016/j.biortech.2018.08.114 (  0) 0) |
| [2] |
Kapoor R, Ghosh P, Kumar M, et al. Valorization of agricultural waste for biogas based circular economy in India: A research outlook. Bioresource Technology, 2020, 304: 123036. DOI:10.1016/j.biortech.2020.123036 (  0) 0) |
| [3] |
Zou J, Liu X, Xu S, et al. Combined hydrothermal pretreatment of agricultural and forestry wastes to enhance anaerobic digestion for methane production. Chemical Engineering Journal, 2024, 486: 150313. DOI:10.1016/j.cej.2024.150313 (  0) 0) |
| [4] |
Xiao L, Lichtfouse E, Kumar P S, et al. Biochar promotes methane production during anaerobic digestion of organic waste. Environmental Chemistry Letters, 2021, 19(5): 3557-3564. DOI:10.1007/s10311-021-01251-6 (  0) 0) |
| [5] |
Bele V, Goyette B, An C, et al. A robust, low-temperature, closed-loop anaerobic system for high-solid mixed farm wastes: Advancing agricultural waste management solutions in canada. Environmental Science and Pollution Research, 2024[2024-06-19]. DOI: 10.1007/s11356-024-33654-7.
(  0) 0) |
| [6] |
Zayen A, Sayadi S, Sousbie P, et al. Chicken manure and wheat straw co-digestion in batch leach bed reactors: Optimization of the start-up conditions. Biomass Conversion and Biorefinery, 2023, 13(12): 10923-10933. DOI:10.1007/s13399-021-01912-0 (  0) 0) |
| [7] |
Chen R, Zhou J, Zheng X, et al. Unveiling the synergy of chlorella sp. and cattle manure co-digestion under high feeding load. Energy, 2023, 270(126877). DOI:10.1016/j.energy.2023.126877 (  0) 0) |
| [8] |
Singh R, Paritosh K, Pareek N, et al. Integrated system of anaerobic digestion and pyrolysis for valorization of agricultural and food waste towards circular bioeconomy: Review. Bioresource Technology, 2022, 360: 127596. DOI:10.1016/j.biortech.2022.127596 (  0) 0) |
| [9] |
Wang Z, Hu Y, Wang S, et al. A critical review on dry anaerobic digestion of organic waste: Characteristics, operational conditions, and improvement strategies. Renewable and Sustainable Energy Reviews, 2023, 176: 113208. DOI:10.1016/j.rser.2023.113208 (  0) 0) |
| [10] |
Weinrich S, Mauky E, Schmidt T, et al. Systematic simplification of the anaerobic digestion model No.1 (ADM1)-laboratory experiments and model application. Bioresource Technology, 2021, 333: 125104. DOI:10.1016/j.biortech.2021.125104 (  0) 0) |
| [11] |
Zhao W, Yang H, He S, et al. A review of biochar in anaerobic digestion to improve biogas production: Performances, mechanisms and economic assessments. Bioresource Technology, 2021, 341: 125797. DOI:10.1016/j.biortech.2021.125797 (  0) 0) |
| [12] |
Ge Y, Tao J, Wang Z, et al. Modification of anaerobic digestion model No.1 with machine learning models towards applicable and accurate simulation of biomass anaerobic digestion. Chemical Engineering Journal, 2023, 454: 140369. DOI:10.1016/j.cej.2022.140369 (  0) 0) |
| [13] |
Emebu S, Pecha J, Janáčová D. Review on anaerobic digestion models: Model classification & elaboration of process phenomena. Renewable and Sustainable Energy Reviews, 2022, 160: 112288. DOI:10.1016/j.rser.2022.112288 (  0) 0) |
| [14] |
Batstone D, Keller J. Industrial applications of the IWA anaerobic digestion model No.1 (ADM1). Water Science and Technology, 2003, 47(1-2): 199-206. DOI:10.2166/wst.2003.0647 (  0) 0) |
| [15] |
Wang L, Li Y, Yi X, et al. Dissimilatory manganese reduction facilitates synergistic cooperation of hydrolysis, acidogenesis, acetogenesis and methanogenesis via promoting microbial interaction during anaerobic digestion of waste activated sludge. Environmental Research, 2023, 218: 114992. DOI:10.1016/j.envres.2022.114992 (  0) 0) |
| [16] |
Mo R, Guo W, Batstone D, et al. Modifications to the anaerobic digestion model No.1 (ADM1) for enhanced understanding and application of the anaerobic treatment processes-A comprehensive review. Water Research, 2023, 244: 120504. DOI:10.1016/j.watres.2023.120504 (  0) 0) |
| [17] |
Tsapekos P, Lovato G, Domingues Rodrigues J A, et al. Amendments to model frameworks to optimize the anaerobic digestion and support the green transition. Renewable and Sustainable Energy Reviews, 2024, 197: 114413. DOI:10.1016/j.rser.2024.114413 (  0) 0) |
| [18] |
Ge Y, Tao J, Wang Z, et al. A hybrid approach of anaerobic digestion model no.1 and machine learning to model and optimize continuous anaerobic digestion processes. Biomass and Bioenergy, 2024, 184: 107176. DOI:10.1016/j.biombioe.2024.107176 (  0) 0) |
| [19] |
Nabaterega R, Nazyab B, Eskicioglu C. Modification and calibration of anaerobic digestion model 1 to simulate volatile fatty acids production during fermentation of municipal sludge. Biochemical Engineering Journal, 2023, 194: 108886. DOI:10.1016/j.bej.2023.108886 (  0) 0) |
| [20] |
Sun H, Yang Z, Shi G, et al. Methane production from acetate, formate and H2/CO2 under high ammonia level: Modified ADM1 simulation and microbial characterization. Science of The Total Environment, 2021, 783: 147581. DOI:10.1016/j.scitotenv.2021.147581 (  0) 0) |
| [21] |
Ashraf R J, Nixon J D, Brusey J. Using multi-objective optimisation with ADM1 and measured data to improve the performance of an existing anaerobic digestion system. Chemosphere, 2022, 301: 134523. DOI:10.1016/j.chemosphere.2022.134523 (  0) 0) |
| [22] |
Trucchia A, Frunzo L. Surrogate based global sensitivity analysis of ADM1-based anaerobic digestion model. Journal of Environmental Management, 2021, 282: 111456. DOI:10.1016/j.jenvman.2020.111456 (  0) 0) |
| [23] |
Alvarado V, Hsu S C, Wu Z, et al. Roadmap from microbial communities to individuality modeling for anaerobic digestion of sewage sludge. Environmental Science & Technology, 2022, 56(10): 6596-6607. DOI:10.1021/acs.est.1c05258 (  0) 0) |
| [24] |
Xie T, Zhang Z, Zhang D, et al. Effect of hydrothermal pretreatment and compound microbial agents on compost maturity and gaseous emissions during aerobic composting of kitchen waste. Science of The Total Environment, 2023, 854: 158712. DOI:10.1016/j.scitotenv.2022.158712 (  0) 0) |
| [25] |
Tampio E, Marttinen S, Rintala J. Liquid fertilizer products from anaerobic digestion of food waste: mass, nutrient and energy balance of four digestate liquid treatment systems. Journal of Cleaner Production, 2016, 125: 22-32. DOI:10.1016/j.jclepro.2016.03.127 (  0) 0) |
| [26] |
Yang Z, Sun H, Zhou L, et al. Bioaugmentation with well-constructed consortia can effectively alleviate ammonia inhibition of practical manure anaerobic digestion. Water Research, 2022, 215: 118244. DOI:10.1016/j.watres.2022.118244 (  0) 0) |
| [27] |
Zhao X, Li L, Wu D, et al. Modified anaerobic digestion model No. 1 for modeling methane production from food waste in batch and semi-continuous anaerobic digestions. Bioresource Technology, 2019, 271: 109-117. DOI:10.1016/j.biortech.2018.09.091 (  0) 0) |
| [28] |
Souza T S O, Carvajal A, Donoso-Bravo A, et al. ADM1 calibration using BMP tests for modeling the effect of autohydrolysis pretreatment on the performance of continuous sludge digesters. Water Research, 2013, 47(9): 3244-3254. DOI:10.1016/j.watres.2013.03.041 (  0) 0) |
| [29] |
de Oliveira Paranhos A G, Adarme O F H, Barreto G F, et al. Methane production by co-digestion of poultry manure and lignocellulosic biomass: Kinetic and energy assessment. Bioresource Technology, 2020, 300: 122588. DOI:10.1016/j.biortech.2019.122588 (  0) 0) |
| [30] |
Chen X, Chen Z, Wang X, et al. Application of ADM1 for modeling of biogas production from anaerobic digestion of Hydrilla verticillata. Bioresource Technology, 2016, 211: 101-107. DOI:10.1016/j.biortech.2016.03.002 (  0) 0) |
| [31] |
Razaviarani V, Buchanan I D. Calibration of the anaerobic digestion model No.1 (ADM1) for steady-state anaerobic co-digestion of municipal wastewater sludge with restaurant grease trap waste. Chemical Engineering Journal, 2015, 266: 91-99. DOI:10.1016/j.cej.2014.12.080 (  0) 0) |
| [32] |
Yan W, Qian T, Soh Y N A, et al. Micro-level evaluation of organic compounds transformation in anaerobic digestion under feast and famine conditions assisted by iron-based materials-revealing the true mechanism of AD enhancement. Environment International, 2020, 135: 105362. DOI:10.1016/j.envint.2019.105362 (  0) 0) |
| [33] |
Białobrzewski I, Waszkielis K, Bułkowska K. The application of anaerobic digestion model No.1 for the optimization of biogas production from maize silage, pig manure, cattle manure, and digestate in a full-scale biogas plant. Fuel, 2024, 35(Part B): 129789. DOI:10.1016/j.fuel.2023.129789 (  0) 0) |
| [34] |
Le T S, Bui X T, Nguyen P D, et al. Bacterial community composition in a two-stage anaerobic membrane bioreactor for co-digestion of food waste and food court wastewater. Bioresource Technology, 2024, 391: 129925. DOI:10.1016/j.biortech.2023.129925 (  0) 0) |
| [35] |
Kong X, Chen J, Wang S, et al. When polyethylene terephthalate microplastics meet Perfluorooctane sulfonate in thermophilic biogas upgrading system: Their effect on methanogenesis. Journal of Hazardous Materials, 2024, 466: 133626. DOI:10.1016/j.jhazmat.2024.133626 (  0) 0) |
| [36] |
Zhang W, Zhang F, Li Y X, et al. No difference in inhibition among free acids of acetate, propionate and butyrate on hydrogenotrophic methanogen of Methanobacterium formicicum. Bioresource Technology, 2019, 294: 122237. DOI:10.1016/j.biortech.2019.122237 (  0) 0) |
| [37] |
Zhang C, Fang Y X, Yin X, et al. The majority of microorganisms in gas hydrate-bearing subseafloor sediments ferment macromolecules. Microbiome, 2023, 11(1): 37. DOI:10.1186/s40168-023-01482-5 (  0) 0) |
| [38] |
Gupta D, Chen K, Elliott S J, et al. MmcA is an electron conduit that facilitates both intracellular and extracellular electron transport in Methanosarcina acetivorans. Nature Communications, 2024, 15(1): 3300. DOI:10.1038/s41467-024-47564-2 (  0) 0) |
| [39] |
Rocha M E, Lazarino T C, Oliveira G, et al. Analysis of biogas production from sewage sludge combining BMP experimental assays and the ADM1 model. PeerJ, 2024, 12: e16720. DOI:10.7717/peerj.16720 (  0) 0) |
| [40] |
Liu J, Xu Y, Wei Y. A model-based approach for evaluating the effects of sludge rheology on methane production during high solid anaerobic digestion: Focusing on mass transfer resistance. Biochemical Engineering Journal, 2024, 201: 109147. DOI:10.1016/j.bej.2023.109147 (  0) 0) |
 2025, Vol. 32
2025, Vol. 32


