2. Department of National Defense Construction Planning and Environmental Engineering, Logistical Engineering University, Chongqing 401311, China
Bioreactors capable of reducing and removing contaminants including ammonia nitrogen, nitrate, and phosphate via microbial metabolism are attracting increasing attention in research and water treatment applications. The microbial community and special functional bacteria, which are the main limiting factors of pollutant degradation, play vital roles in bioreactors[1]. Recent years have witnessed great efforts focused on investigating the impact of influent condition heterogeneity, especially changes in substrate concentration, on bacterial growth, bacterial community composition[2-3], and ultimately microbial diversity[4-5], as well as on water treatment efficiency at high substrate concentrations[6]. These studies have revealed that the speed of bacterial response to substrate has been relatively slow at high substrate concentration, with tens of days often being required for the microbial community to adapt to influent heterogeneity.
Most studies conducted to date have investigated high substrate concentrations. However, few investigations have evaluated the impacts of influent heterogeneity on bacterial community diversity and biotreatment performance of micro-polluted source water at low concentrations of organic carbon and ammonia nitrogen[7]. Given that low nutrient concentrations restrict microbial growth[8] and render the bacterial community more susceptible to environmental changes[9], concerns regarding the influence of influent conditions at low nutrient levels on the bacterial community and effluent quality are needed to improve treatment of drinking water, as well as the design and start-up of waterworks.
Polymerase chain reaction-denaturing gradient gel electrophoresis (PCR-DGGE) has been widely applied for biodiversity assessment in bioreactor samples and drinking water treatment[10] to identify members and dominant bacteria of the microbial community in bioreactors. The Shannon diversity index (H) based on DGGE analysis is regularly employed in the analysis of bacterial diversity[11]. Additionally, cluster analysis of the data obtained from PCR-DGGE is helpful to understand the ecological status of a bioreactor system and determine key factors influencing water quality[12].
The aim of this investigation was two-fold: 1) to assess the operation characteristics of different bioreactor systems that shape structures and functions of the bacterial community during treatment processes, such as influent organic concentration, influent ammonia nitrogen concentration and hydraulic retention time (HRT); 2) to gain insight into the diversity of the bacterial community induced from environmental adaptability in parallel operations based on DGGE and cluster analyses. The results of the present study suggested that influent heterogeneity plays a significant role in structuring the bacterial community, and higher influent organic concentrations promote enrichment of species during the setup of drinking water treatment processes.
2 Experiment Methods 2.1 Experimental Setup and OperationThe experiments were conducted in nine plexiglass laboratory-scale biological aerated filter (BAF) reactors operated in parallel. A schematic diagram of a BAF is provided in Fig. 1. Cylindrical reactors (550 mm height, 55 mm inner diameter) with an effective volume of 1 L were run in a continuous flow. The peristaltic pumps were actuated to control the HRT and gas valves were adjusted to regulate the aeration rate to maintain the dissolved oxygen (DO) concentration at 6-7 mg/L. The biomass was attached to suspended carriers filled in a volume fraction of 30%-40%.
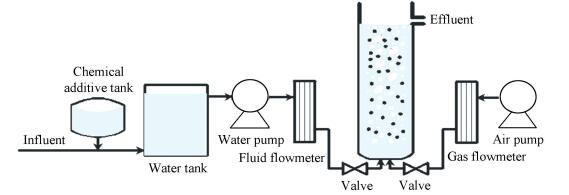
|
Figure 1 Schematic diagram of the experimental setup |
The Pearl River system in Guangdong Province was supplied as raw water with an average temperature of 27 ℃. Reactors were fed with ammonium chloride and sodium acetate to simulate various influent qualities. The operating conditions of different reactors are provided in Table 1. For experimental runs 1 to 3, the influencing factor was influent NH3-N concentration, for experimental runs 4 to 6, the influencing factor was HRT and for experimental runs 7 to 9, the primary influencing factor of ammonia nitrogen removal rate was organic matter.
| Table 1 Experimental conditions of BAF |
2.2 Analytical Methods
All samples were filtered through 0.45 μm cellulose acetate membrane before analysis. The water quality parameters measured included NH3-N, CODMn, UV254, DO and pH. The NH3-N and CODMn concentrations were determined according to the ammonium-Nessler's reagent colorimetric and potassium permanganate method, respectively[13-14]. The DO and pH were measured using a dissolved oxygen meter (DO200, YSI, USA) and a pH electrode (PHS-3C, Rex, China).
2.3 Scanning Electron Microscopy (SEM) AnalysisThe shape structures and distribution of the bacterial community on the surfaces of suspended carriers were characterised by SEM (Carl Zeiss EV0 LS10, Germany) analysis. Sequential procedures of sample pretreatments included fixation with 2.5% glutaraldehyde (Volume ratio, 4 ℃, 1.5 h), dehydration using graded ethanol (50%, 70%, 90%, 100%, volume ratio, room temperature, 1.0 h), drying, and gold spraying using a precision etching coating system (Gatan Model 682).
2.4 DNA ExtractionBiofilm samples collected from suspended carriers in each reactor were centrifuged to provide at least 0.25 g total biomass. Subsequently, DNA was isolated using a PowerSoil DNA Isolation Kit (Mo Bio Laboratories, Inc., Carlsbad, CA, USA) according to the manufacturer's instructions.
Amplification of 16S rRNA for DGGE analysis by polymerase chain reaction (PCR) on a Mastercycler Gradient Thermalcycler (Eppendorf, Germany) was conducted using the general bacterial primers 343F-GC and 534R (343F, 5'-ACTCCTACGGGAGGCAGCAG-3' and 534R, 5'-ATTACCGCGGCTGCTGG-3' with a GC clamp 5'-CGCCCGCCGCGCGCGGCGGGCGGGGCGGGGGCCCGGGGG-3'). PCR was conducted by subjecting the samples to the following conditions: preheating at 94 ℃ for 10 min, followed by 30 cycles of denaturation at 94 ℃ for 1 min, annealing at 55 ℃ for 30 s and elongation at 72 ℃ for 90 s, and then final extension at 72 ℃ for 10 min, after which the samples were held at 4 ℃. The reaction system consisted of 0.25 μL Taq DNA polymerase (Takara, Dalian, China), 5 μL PCR Buffer (Mg2+ Plus), 4 μL dNTP Mixture (2.5 mmol/L), 2.5 ng DNA template, 1 μL 343F primer (20 μmol/L), 1 μL 534R primer (20 μmol/L), and 35.75 μL Milli-Q water. Following amplification, samples were subjected to 1% agarose gel electrophoresis to verify the presence of PCR products.
DGGE was conducted using a DcodeTM universal mutation detection system (BioRad Laboratories, Hercules, CA, USA). An 8% polyacrylamide gel with a 30%-60% denaturant gradient (100% denaturant comprises 7 mol/L urea and 40% deionised formamide) was loaded by adding 10 μL of PCR amplification products and 2 μL of loading buffer, after which samples were electrophoresed at 150 V and 60 ℃ for 7 h. Finally, the gels were silver stained and scanned with a transmission scanner.
The target DNA bands were excised from the gels and dissolved in 30 μL TE. Next, the PCR products were purified using a DNA Extraction Kit (D823A, Takara, Japan), after which the target DNA was connected to a pMD 18-T Vector (D101A, Takara, Japan). Bacterium solution PCR was then performed using the M13 universal primers, after which the samples were sequenced using an ABI 3730XL DNA Analyzer (Applied Biosystems, Foster City, CA, USA). Finally, the sequences were subjected to a BLAST search (NCBI, US National Library of Medicine) for species identification.
2.5 Statistical AnalysisThe scanned gels containing DNA band profiles were analysed to determine the microbial diversity using molecular biological software. The Shannon-Weaver index of species diversity (H) was then calculated according to
| $ H = -\sum\limits_{i = 0}^t {{P_I}\ln \left( {{P_i}} \right)} $ | (1) |
where Pi is the proportion of band i in a DGGE profile, and t is the total number of bands. The Shannon-Weaver index, which is a measurement of both numbers and proportions of species, is particularly useful in identifying the abundance of bacterial communities[15].
After removal of the background, the clustering algorithm of the unweighted pair group method of arithmetic mean (UPGMA) was used to calculate dendrograms[16] and obtain the Dice coefficient similarity coefficient (SD), which represented similarities between pairs of samples[17]. The microbial resolution in this study, which represented the differences between pairs of samples, was calculated using the equation: 1-SD.
3 Results and Discussion 3.1 Effects of Influent Conditions on Surface MorphologyAll nine suspended carrier bioreactors went through a start-up period in which the microbial community was gradually formed and a high ammonia nitrogen removal rate was achieved. Scanning electron microscopy (SEM) was conducted to investigate bacterial morphology of biofilm on plastic carriers in varied bioreactors. As shown in Fig. 2, an extensive survey of microscopic features revealed several areas where rod-shaped bacteria marked with white arrows had apparently occurred. Long bacilli and coccoid-shaped bacteria appeared and coexisted with the typical rod-shaped bacteria with increasing concentration of ammonia nitrogen (Fig. 2(a) to 2(c)). The filamentous formations or some specific extracellular structures instead of microorganisms occurred on the superficial layers of biofilms. Short HRT may be responsible for the phenomenon because such extracellular structures are frequently observed on the surfaces of bacteria living under conditions of fast-flowing stream[18]. Moreover, extension of HRT promoted aggregation of biofilm networks (Fig. 2(d) to 2(f)). It was remarkable the typical rod-shaped bacteria appeared again in Fig. 2(h), meanwhile abundant filamentous structures transformed into dense biofilm with increasing concentration of organic matter (Fig. 2(g) to 2(i)). It could be seen that typical rod-shaped bacteria which marked with white arrows in SEM images inserted or spread on the surfaces of the biofilms.
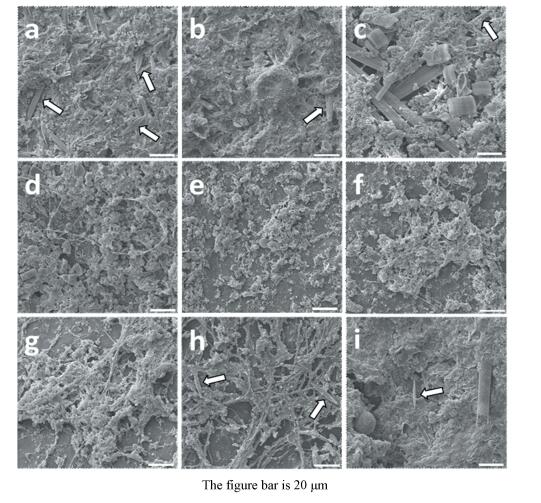
|
Figure 2 SEM images of the microstructure of suspended carriers surfaces at different experimental runs of all BAF systems |
According to Bergey's Manual of Systematic Bacteriology[19], nitrifying bacteria are not characterised by filamentous bacteria, such as Nitrobacter, Nitrospira, Nitrosomonas and Nitrosococcus, thus, the results of the present study indicate that enhancement of organic matter promotes the growth of heterotrophic bacteria. These observations are in agreement with the results of previous studies[20]. Ammonium availability and influent HRT also shaped the community structures[21]. In the present study, both increasing ammonia nitrogen concentration and extension of HRT also led to morphological heterogeneity.
3.2 Effects of Influent Conditions on Bacterial DiversityThe DGGE band analysis based on Shannon-Weaver index (H), which indicates microbial community diversity, has been used to characterise changes in community structure[22]. A high H value indicates high species diversity, and our research showed that the H value differed across influent conditions (Fig. 3). In experimental runs 1, 2 and 3 (Table 1), when the influent organic carbon and HRT were maintained at the same levels, the H values increased from 2.078 to 2.395 and 2.479 (runs 1-3) as the ammonia nitrogen concentration increased (Fig. 3). The diversity index was also impacted by the HRT. Specifically, the H values were 2.479, 2.635 and 2.769, respectively, in the following three bioreactors (runs 4-6), which had increasing HRT values. As the concentration of the influent organic carbon gradually increased, the H values first decreased from 2.636 to 2.501, then increased to 2.633 (runs 7-9). Previous study demonstrated that increasing of organic matter contents could promote the growth of nitrifying bacteria within a certain concentration range[23], and then the nitrifying bacteria which belong to chemoautotrophs competed with heterotrophs. Thus, decreasing of H value in run 8 could be due to reducing of heterotrophs in bacterial community. However, persistent increasing of organic matter concentration finally brought about growth advantage of heterotrophs that helped H value recover in run 9.
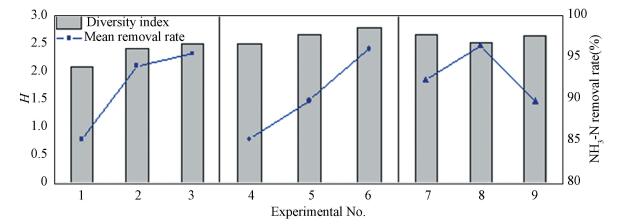
|
Figure 3 Bacterial species diversity using the Shannon-Weaver index (H) and NH3-N average removal efficiency for each bioreactor |
The differences in the diversity indices in the various bioreactors mentioned above were small compared with those reported in previous studies[24-25], which was likely due to the low concentration of substrates and relatively short HRT in our system.
A previous study revealed that the initial substrate intensely affected microbial diversity[26]; therefore, it is important to consider available nutrient sources[27] when analysing the H results. In our bioreactor systems, ammonia nitrogen and organic matter, which served as nitrogen and carbon sources, provided nourishment for autotrophic and heterotrophic bacteria, respectively. The increase in H values was positively correlated with increasing of ammonia nitrogen concentration and extended HRT, and prolonging the HRT was equivalent to reducing the organic carbon concentrations and influent organic loading. However, there was no apparent variation tendency when the concentrations of organic matter and ammonia nitrogen increased simultaneously. These results demonstrate that alterations in H value mainly stemmed from changes in ammonia nitrogen concentration, and thatthe influent organic matter had a negative effect on H values. Moreover, as shown in Fig. 3, the variations in average NH3-N removal efficiency were in agreement with the diversity index in the first six bioreactors. For experimental runs 7 to 9, there were completely opposite trends between H values and removal rates. The trends illustrated that high levels of organic matter weakened the removal efficiency of ammonia nitrogen and damaged the stability of the nitrifying bacterial communities because of enhanced competition among bacterial species.
3.3 Effects of Influent Conditions on Dominant SpeciesThe DGGE profiles generated from bioreactors of different influent conditions demonstrated that the NH3-N concentration, organic carbon concentration and HRT shaped the construction of flora and led to the unique bacterial community (Fig. 4).
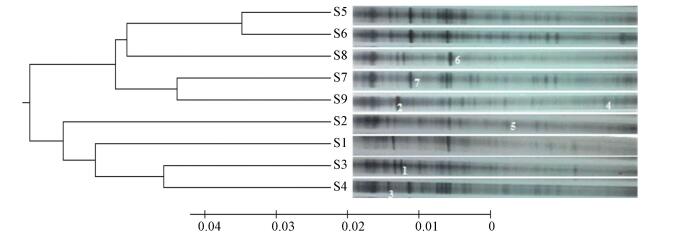
|
Figure 4 Bacterial community DGGE profiles and UPGMA tree |
Cluster analysis using the UPGMA algorithm indicated that the principal factor brought about the separation of the community was the organic carbon level. The continuous increase in CODMn level divided the species into two main parts, and the influent NH3-N level caused a further divergence. The results of microbial diversity and cluster analysis showed that increasing organic carbon concentrations would lead to changes in bacterial composition, but not enhancement of H values. Furthermore, UPGMA clustering based on the Dice similarity coefficient (Table 2) of the DGGE fingerprints showed a large difference between run 1 and run 9, and the microbial resolution primarily generated from different concentrations of CODMn reached 0.111. Another high value of resolution occurred between run 1 and run 6 in response to minor alteration of the HRT. DGGE fingerprinting indicated that bacterial communities responded sensitively to influent quality and hydraulic condition.
| Table 2 Comparison of microbial resolution with influent conditions |
Sequences of approximately 200 nucleotides corresponding to region V3 were ultimately acquired from nine DGGE bands, and the identified bands dominated after the stable operation of bioreactors. The species identification results (Table 3) implied that nitrifying bacteria including Nitrobacter winogradskyi, Candidatus Nitrospira, Nitrosomonas nitrosa and Nitrospira moscoviensis played a leading role in the entire group of bacteria, which could explain the high efficiency of NH3-N removal for each bioreactor and the negative influence of influent organic carbon concentration for H values. Interestingly, run 3 and run 8 both contained a band corresponding to Nitrobacter winogradskyi while run 7 and run 9 did not have the same pattern as run 8, and instead contained Sphingomonas wittichii and Chryseolinea serpens serving as the dominant bacteria respectively.
| Table 3 Similar sequence of DGGE bands of relatively dominant bacteria |
The species identification results (Table 3) showed that nitrifying bacteria were dominant in each bioreactor to ensure a high removal rate of NH3-N, however, influent conditions also led to differences in the bacterial communities. Specifically, sample 1, sample 2 and sample 3 shared similar bands, as shown in Fig. 4, and only the types of nitrifying bacteria differed. Hence, the influent NH3-N concentration mainly changed the types of autotrophic bacteria. Comparison of samples 4-6, in which the HRTs were different, indicated there were no obvious distinctions in communities. Finally, increasing concentrations of influent organic matter led to remarkable changes in bacterial types. The dominant bacteria in samples 7-9 were Sphingomonas wittichii, Nitrobacter winogradskyi and Chryseolinea serpens, respectively.Nitrobacter winogradskyi are autotrophic bacteria, while Sphingomonas wittichii and Chryseolinea serpens are heterotrophic bacteria.
High resolution value between runs 1 and 9 that their difference of substrate concentration was the greatest revealed large distinctions of bacterial communities as shown in Table 2. Moreover, autotrophs and heterotrophs were dominant in run 1 and run 9, respectively (Fig. 4). These results demonstrated varied concentrations of substrates influenced compositions of bacterial communities, and then the competition occurred between autotrophic and heterotrophic bacteria. Previous studies also reported that competition between the two types of bacteria mainly originated from changes in organic substrate loading[28]. The results also suggested that competition among different heterotrophic bacteria weakened the NH3-N removal efficiency.
4 ConclusionsAt low substrate concentration, influent heterogeneity impacted bacterial community diversity, degree of microbial resolution and effluent quality. The influence was relatively weaker than that at high substrate concentration, but each bioreactor system fed back sensitively. SEM and PCR-DGGE confirmed that different conditions led to diverse morphological characteristics and distributions in dominant bacteria. Cluster analysis showed that influent CODMn concentration was the key factor influencing bacterial succession. Molecular biological analysis, revealed responses of the bacterial community to micro-polluted source water and effects of CODMn on NH3-N removal efficiency. In addition, nitrifying bacteria were active as the dominant bacteria.
| [1] |
Sousa J A B, Plugge C M, Stams A J M, et al. Sulfate reduction in a hydrogen fed bioreactor operated at haloalkaline conditions. Water Research, 2015, 68: 67-76. DOI:10.1016/j.watres.2014.09.035 ( 0) 0)
|
| [2] |
Griffiths B S, Ritz K, Ebblewhite N, et al. Soil microbial community structure: effects of substrate loading rates. Soil Biology and Biochemistry, 1999, 31: 145-153. ( 0) 0)
|
| [3] |
Palatsi J, Viñas M, Guivernau M, et al. Anaerobic digestion of slaughterhouse waste: Main process limitations and microbial community interactions. Bioresource Technology, 2011, 102: 2219-2227. DOI:10.1016/j.biortech.2010.09.121 ( 0) 0)
|
| [4] |
Degens B P, Harris J A. Development of a physiological approach to measuring the catabolic diversity of soil microbial communities. Soil Biology and Biochemistry, 1997, 29: 1309-1320. DOI:10.1016/S0038-0717(97)00076-X ( 0) 0)
|
| [5] |
Nannipieri P, Ascher J, Ceccherini M T, et al. Microbial diversity and soil functions. European Journal of Soil Science, 2003, 54: 655-670. DOI:10.1046/j.1351-0754.2003.0556.x ( 0) 0)
|
| [6] |
Fan J L, Wang W G, Zhang B, et al. Nitrogen removal in intermittently aerated vertical flow constructed wetlands: Impact of influent COD/N ratios. Bioresource Technology, 2013, 143: 461-466. DOI:10.1016/j.biortech.2013.06.038 ( 0) 0)
|
| [7] |
Chu H Q, Dong B Z, Zhang Y L, et al. Pollutant removal mechanisms in a bio-diatomite dynamic membrane reactor for micro-polluted surface water purification. Desalination, 2012, 293: 38-45. DOI:10.1016/j.desal.2012.02.021 ( 0) 0)
|
| [8] |
Egli T. How to live at very low substrate concentration. Water Research, 2010, 44: 4826-4837. DOI:10.1016/j.watres.2010.07.023 ( 0) 0)
|
| [9] |
Lautenschlager K, Hwang C C, Ling F Q, et al. Abundance and composition of indigenous bacterial communities in a multi-step biofiltration-based drinking water treatment plant. Water Research, 2014, 62: 40-52. DOI:10.1016/j.watres.2014.05.035 ( 0) 0)
|
| [10] |
Araujo J C, Schneider R P. DGGE with genomic DNA: Suitable for detection of numerically important organisms but not for identification of the most abundant organisms. Water Research, 2008, 42: 5002-5010. DOI:10.1016/j.watres.2008.09.011 ( 0) 0)
|
| [11] |
Miura Y K, Hiraiwa M N, Ito T, et al. Bacterial community structures in MBRs treating municipal wastewater: Relationship between community stability and reactor performance. Water Research, 2007, 41: 627-637. DOI:10.1016/j.watres.2006.11.005 ( 0) 0)
|
| [12] |
Reghunath R, Murthy T R S, Raghavan B R. The utility of multivariate statistical techniques in hydrogeochemical studies: an example from Karnataka, India. Water Research, 2002, 36: 2437-2442. DOI:10.1016/S0043-1354(01)00490-0 ( 0) 0)
|
| [13] |
Zhang Z H, Yuan Y, Fang Y J, et al. Preparation of photocatalytic nano-ZnO/TiO2 film and application for determination of chemical oxygen demand. Talanta, 2007, 73(3): 523-528. DOI:10.1016/j.talanta.2007.04.011 ( 0) 0)
|
| [14] |
Qin W, Li W G, Zhang D Y, et al. Ammonium removal of drinking water at low temperature by activated carbon filter biologically enhanced with heterotrophic nitrifying bacteria. Environmental Science and Pollution Research, 2016, 23: 4650-4659. DOI:10.1007/s11356-015-5561-9 ( 0) 0)
|
| [15] |
Li A J, Yang S F, Li X Y, et al. Microbial population dynamics during aerobic sludge granulation at different organic loading rates. Water Research, 2008, 42: 3552-3560. DOI:10.1016/j.watres.2008.05.005 ( 0) 0)
|
| [16] |
Lautenschlager K, Hwang C C, Liu W T, et al. A microbiology-based multi-parametric approach towards assessing biological stability in drinking water distribution networks. Water Research, 2013, 47: 3015-3025. DOI:10.1016/j.watres.2013.03.002 ( 0) 0)
|
| [17] |
Yan Q Y, Yu Y H, Feng W S, et al. Genetic diversity of plankton community as depicted by PCR-DGGE fingerprinting and its relation to morphological composition and environmental factors in Lake Donghu. Microbial Ecology, 2007, 54: 290-297. DOI:10.1007/s00248-006-9200-3 ( 0) 0)
|
| [18] |
Ridgway H F, Olson B H. Scanning electron microscope evidence for bacterial colonization of a drinking-water distribution system. Applied and Environmental Microbiology, 1981, 41: 274-287. ( 0) 0)
|
| [19] |
Krieg N R, Holt J G. Bergey's Manual of Systematic Bacteriology. Baltimore, USA: Williams and Wilkins, 1984.
( 0) 0)
|
| [20] |
Jackson C R. Changes in community properties during microbial succession. Oikos, 2003, 101: 444-448. DOI:10.1034/j.1600-0706.2003.12254.x ( 0) 0)
|
| [21] |
Mouser P J, N' Guessan A L, Elifantz H, et al. Influence of heterogeneous ammonium availability on bacterial community structure and the expression of nitrogen fixation and ammonium transporter genes during in situ bioremediation of uranium-contaminated groundwater. Environmental Science and Technology, 2009, 43: 4386-4392. DOI:10.1021/es8031055 ( 0) 0)
|
| [22] |
Shannon C E, Weaver W. The Mathematical Theory of Communication. Urbana, IL: UIllinois Press, 1949.
( 0) 0)
|
| [23] |
Wang X P, Chen J, Hua Z Z, et al. The optimum growth and nitrification conditions of nitrifying bacteria. Chinese Journal Applied Environmental Biology, 1995, 5(1): 64-68. ( 0) 0)
|
| [24] |
Krakat N, Schmidt S, Scherer P. Mesophilic fermentation of renewable biomass: Does hydraulic retention time regulate methanogen diversity? Applied and Environmental Microbiology, 2010, 76: 6322-6326. Applied and Environmental Microbiology, 2010, 43: 6322-6326. ( 0) 0)
|
| [25] |
Adrados B, Sánchez O, Arias C A, et al. Microbial communities from different types of natural wastewater treatment systems: Vertical and horizontal flow constructed wetlands and biofilters. Water Research, 2014, 55: 304-312. DOI:10.1016/j.watres.2014.02.011 ( 0) 0)
|
| [26] |
Alidina M, Li D, Drewes J E. Investigating the role for adaptation of the microbial community to transform trace organic chemicals during managed aquifer recharge. Water Research, 2014, 56: 172-180. DOI:10.1016/j.watres.2014.02.046 ( 0) 0)
|
| [27] |
Jones B E, Grant W D, Duckworth A W, et al. Microbial diversity of soda lakes. Extremophiles, 1998, 2: 191-200. DOI:10.1007/s007920050060 ( 0) 0)
|
| [28] |
Elenter D, Milferstedt K, Zhang W, et al. Influence of detachment on substrate removal and microbial ecology in a heterotrophic/autotrophic biofilm. Water Research, 2007, 41: 4657-4671. DOI:10.1016/j.watres.2007.06.050 ( 0) 0)
|
 2017, Vol. 24
2017, Vol. 24


